使用发布的代码 在这里,我创建了一个很好的层次聚类:
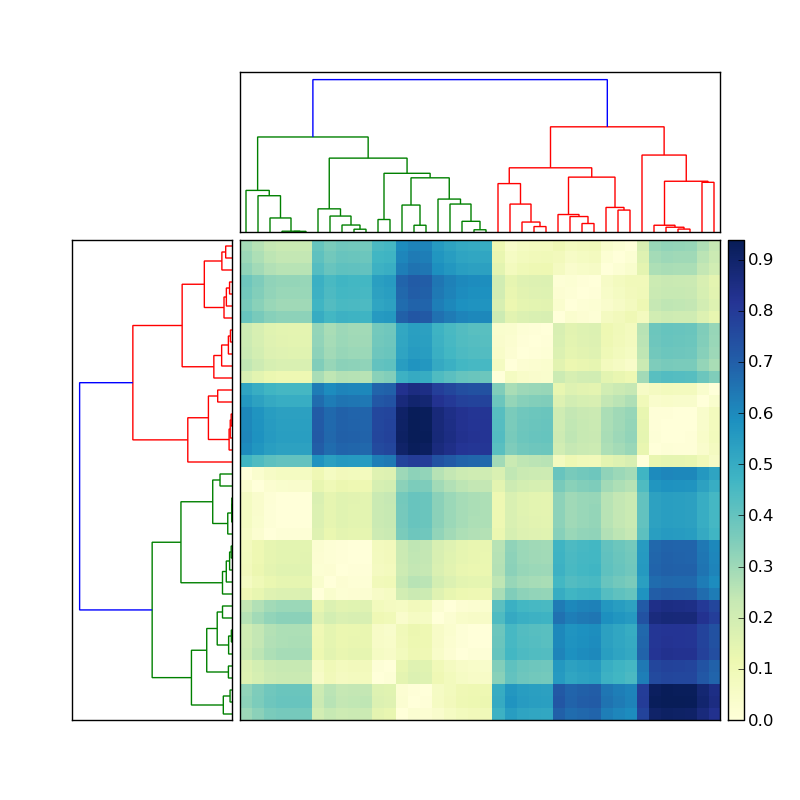
假设左侧的树状图是通过执行以下操作创建的
Y = sch.linkage(D, method='average') # D is a distance matrix
cutoff = 0.5*max(Y[:,2])
Z = sch.dendrogram(Y, orientation='right', color_threshold=cutoff)
:现在如何获取每个彩色簇的成员的索引? 为了简化这种情况,忽略顶部的聚类,仅关注矩阵左侧的树状图。
此信息应存储在树状图 Z 存储变量中。有一个函数应该可以做我想要的事情,称为fcluster(请参阅文档此处)。但是,我看不到在哪里可以为 fcluster 提供与我在创建树状图时指定的相同的 cutoff。看来 fcluster 中的阈值变量,t 必须根据各种模糊的测量值(不一致不一致,距离>、maxclust、monocrit)。有什么想法吗?
Using the code posted here, I created a nice hierarchical clustering:

Let's say the the dendrogram on the left was created by doing something like
Y = sch.linkage(D, method='average') # D is a distance matrix
cutoff = 0.5*max(Y[:,2])
Z = sch.dendrogram(Y, orientation='right', color_threshold=cutoff)
Now how do I get the indices of the members of each of the colored clusters? To simplify this situation, ignore the clustering on the top, and focus only on the dendrogram on the left of the matrix.
This information should be stored in the dendrogram Z stored variable. There is a function that should do just what I want called fcluster (see documentation here). However I cannot see where I can give fcluster the same cutoff as I specified in the creation of the dendrogram. It seems that the threshold variable in fcluster, t has to be in terms of various obscure measurements (inconsistent, distance, maxclust, monocrit). Any ideas?




发布评论
评论(4)
我认为你走在正确的道路上。让我们试试这个:
ind将为您提供 100 个输入观测值中每个观测值的聚类索引。ind取决于您在linkage中使用的方法。尝试method=single、complete和average。然后注意ind有何不同。示例:
scipy.cluster.hierarchy确实令人困惑。在你的链接中,我什至不认识我自己的代码!I think you're on the right track. Let's try this:
indwill give you cluster indices for each of the 100 input observations.inddepends on whatmethodyou used inlinkage. Trymethod=single,complete, andaverage. Then note howinddiffers.Example:
scipy.cluster.hierarchysure is confusing. In your link, I don't even recognize my own code!我编写了一些代码来解压缩链接矩阵。它返回一个字典,其中包含按每个聚合步骤分组的标签索引。我只在
complete链接集群的结果上进行了尝试。字典的键从 len(labels)+1 开始,因为最初,每个标签都被视为自己的簇。这可能会回答你的问题。返回:
I wrote some code to decondense the linkage matrix. It returns a dictionary containing the indexes of
labelsthat are grouped by each agglomeration step. I've only tried it out on the results of thecompletelinkage clusters. The keys of the dict start atlen(labels)+1because initially, each label is treated as its own cluster. This may answer your question.Returns:
我知道这对游戏来说已经很晚了,但是我根据帖子 此处。它已在 pip 上注册,因此要安装,您只需调用
此处查看该项目的 github 页面:https://github。 com/themantalope/pydendroheatmap
I know this is very late to the game, but I made a plotting object based on the code from the post here. It's registered on pip, so to install you just have to call
check out the project's github page here : https://github.com/themantalope/pydendroheatmap
您还可以尝试
cut_tree,它有一个高度参数,可以为您提供超测量所需的信息。You can also try
cut_tree, it has a height parameter that should give you what you want for ultrametrics.