绘制加权频率矩阵
这个问题与我之前问过的两个不同问题有关:
1)再现频率矩阵图
2 ) 向累积图添加 95% 置信限
I希望在 R 中重现此图: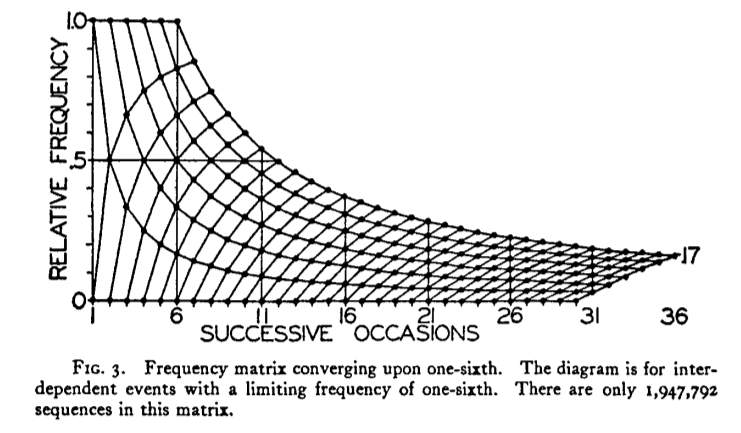
我已经使用图形下面的代码: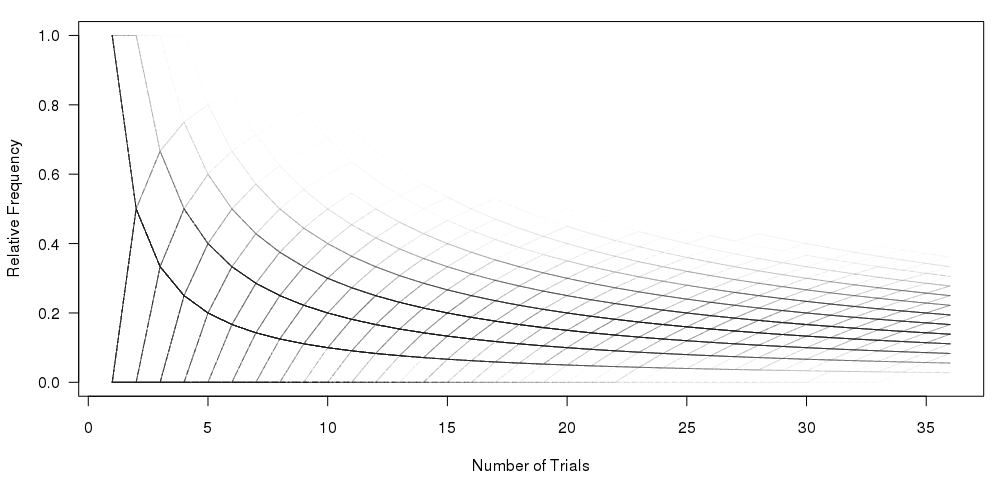
#Set the number of bets and number of trials and % lines
numbet <- 36
numtri <- 1000
#Fill a matrix where the rows are the cumulative bets and the columns are the trials
xcum <- matrix(NA, nrow=numbet, ncol=numtri)
for (i in 1:numtri) {
x <- sample(c(0,1), numbet, prob=c(5/6,1/6), replace = TRUE)
xcum[,i] <- cumsum(x)/(1:numbet)
}
#Plot the trials as transparent lines so you can see the build up
matplot(xcum, type="l", xlab="Number of Trials", ylab="Relative Frequency", main="", col=rgb(0.01, 0.01, 0.01, 0.02), las=1)
我的问题是:如何在不绘制多个样本的情况下一次性重现顶部图?
谢谢。
如果你对这篇内容有疑问,欢迎到本站社区发帖提问 参与讨论,获取更多帮助,或者扫码二维码加入 Web 技术交流群。

绑定邮箱获取回复消息
由于您还没有绑定你的真实邮箱,如果其他用户或者作者回复了您的评论,将不能在第一时间通知您!


发布评论
评论(3)
生成此图...
您可以使用以下代码
You can produce this plot...
... by using this code:
您还可以使用 Koshke 的方法,通过将值的组合限制为 s<6 的值,并根据 Andrie 的要求添加 Ps$n 和 ps$s 差异的条件,以获得“指向的”配置。
You can also use Koshke's method, by limiting the combinations of values to those with s<6 and at Andrie's request added the condition on the difference of Ps$n and ps$s to get a "pointed" configuration.
加权频率矩阵也称为位置权重矩阵(生物信息学)。
它可以以序列徽标的形式表示。
这至少是我绘制加权频率矩阵的方式。
Weighted Frequency Matrix is also called Position Weight Matrix (in bioinformatics).
It can be represented in a form of a sequence logo.
This is at least how I plot weighted frequency matrix.