使用 ggplot2 再现格子树状图
这可以用 ggplot2 重现这个格子图吗?
library(latticeExtra)
data(mtcars)
x <- t(as.matrix(scale(mtcars)))
dd.row <- as.dendrogram(hclust(dist(x)))
row.ord <- order.dendrogram(dd.row)
dd.col <- as.dendrogram(hclust(dist(t(x))))
col.ord <- order.dendrogram(dd.col)
library(lattice)
levelplot(x[row.ord, col.ord],
aspect = "fill",
scales = list(x = list(rot = 90)),
colorkey = list(space = "left"),
legend =
list(right =
list(fun = dendrogramGrob,
args =
list(x = dd.col, ord = col.ord,
side = "right",
size = 10)),
top =
list(fun = dendrogramGrob,
args =
list(x = dd.row,
side = "top",
size = 10))))

Is this possible to reproduce this lattice plot with ggplot2?
library(latticeExtra)
data(mtcars)
x <- t(as.matrix(scale(mtcars)))
dd.row <- as.dendrogram(hclust(dist(x)))
row.ord <- order.dendrogram(dd.row)
dd.col <- as.dendrogram(hclust(dist(t(x))))
col.ord <- order.dendrogram(dd.col)
library(lattice)
levelplot(x[row.ord, col.ord],
aspect = "fill",
scales = list(x = list(rot = 90)),
colorkey = list(space = "left"),
legend =
list(right =
list(fun = dendrogramGrob,
args =
list(x = dd.col, ord = col.ord,
side = "right",
size = 10)),
top =
list(fun = dendrogramGrob,
args =
list(x = dd.row,
side = "top",
size = 10))))

如果你对这篇内容有疑问,欢迎到本站社区发帖提问 参与讨论,获取更多帮助,或者扫码二维码加入 Web 技术交流群。

绑定邮箱获取回复消息
由于您还没有绑定你的真实邮箱,如果其他用户或者作者回复了您的评论,将不能在第一时间通知您!

发布评论
评论(5)
正如本所说,一切皆有可能。一些支持树状图的工作已经完成。 Andrie de Vries 提出了一种强化方法树对象。然而,生成的图形并不像您所看到的那样漂亮。
瓷砖很容易做。对于树状图,我将检查
plot.dendrogram(使用getAnywhere)以查看如何计算线段的坐标。提取这些坐标并使用 geom_segment 绘制树状图。然后使用视口将图块和树状图一起绘制。抱歉,我无法举例,这需要大量工作,而且为时已晚。我希望这有帮助
干杯
As Ben says, everything is possible. Some work to support dendrograms has been done. Andrie de Vries has made a fortify method of tree objects. However, the resulting graphic is not pretty as you can see.
The tile would be easy to do. For the dendrogram I would inspect
plot.dendrogram(usinggetAnywhere) to see how the coordinates for the segments are calculated. Extract those coordinates and use geom_segment to plot the dendrogram. Then use viewports to plot the tiles and the dendrogram together. Sorry I can't give a example, it's a lot of work and it's too late.I hope this helps
Cheers
疑。我在 ggplot2 索引中没有看到任何建议支持树状图的函数,当这位博主将 Sarkar 的 Lattice 书中插图的一组翻译放在一起时,他无法获得 ggplot 树状图图例:
http://learnr.wordpress.com/2009/08/10/ggplot2-version-of-figures-in-lattice-multivariate-data-visualization-with-r-part-9/
Doubtful. I do not see any functions in the Index for ggplot2 that would suggest support for dendrograms, and when this blogger put together a set of translations of the illustrations in Sarkar's Lattice book, he was unable to get a ggplot dendrogram legend:
http://learnr.wordpress.com/2009/08/10/ggplot2-version-of-figures-in-lattice-multivariate-data-visualization-with-r-part-9/
这些链接为 ggplot2 中带有树状图的热图提供了解决方案:
https://gist.github.com/chr1swallace/ 4672065
https://github.com/chr1swallace/random-functions/ blob/master/R/ggplot-heatmap.R
还有这个:
垂直对齐 ggplot2 绘图
These links provide a solution for heatmaps with dendrograms in ggplot2:
https://gist.github.com/chr1swallace/4672065
https://github.com/chr1swallace/random-functions/blob/master/R/ggplot-heatmap.R
and also this one:
Align ggplot2 plots vertically
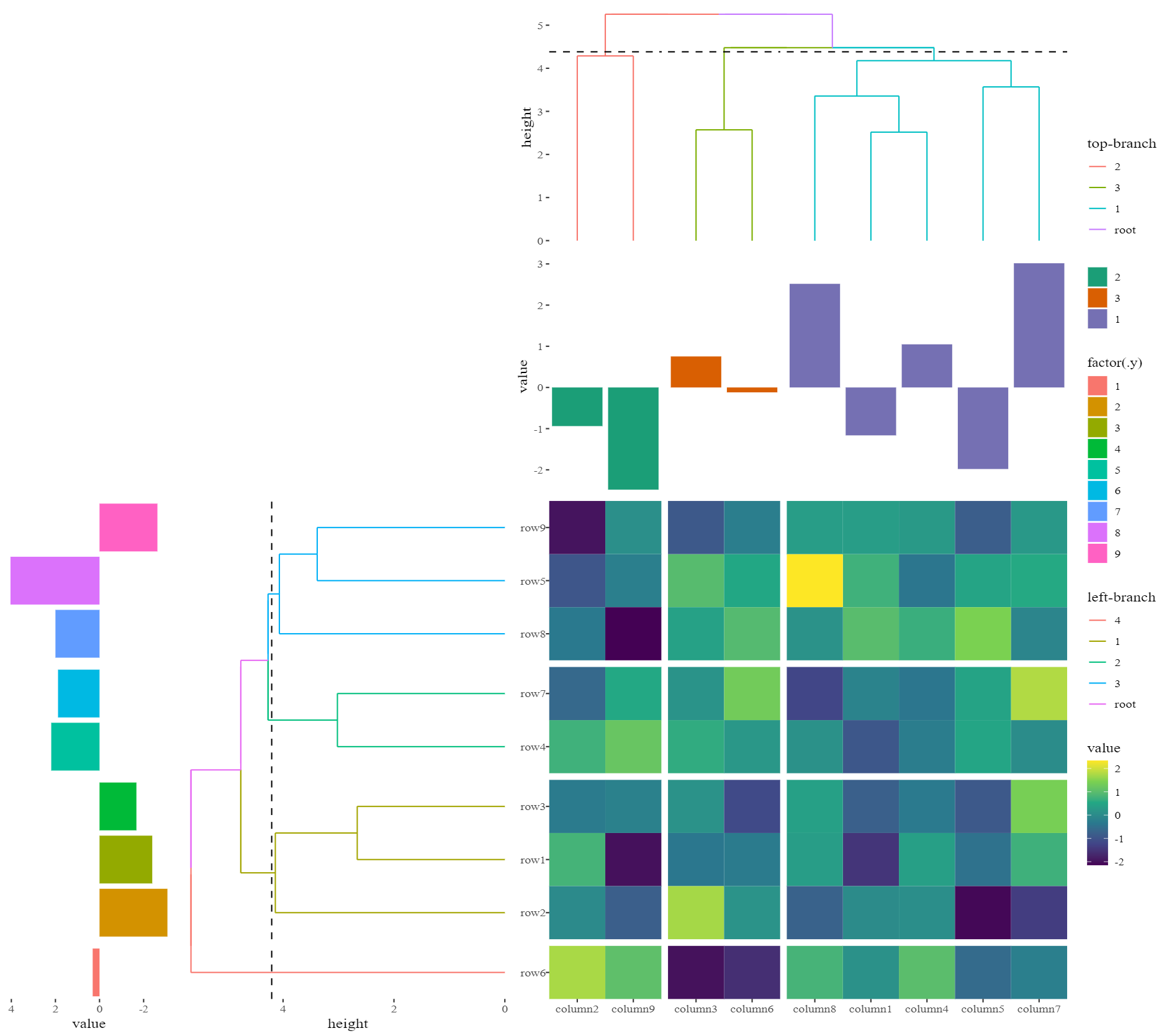
如果您想使用 ggplot2 来创建带有树状图的热图。我创建了一个名为 ggalign 的包,您可以使用它来创建与
一样复杂的热图Complexheatmap:具体来说,它可以将热图拆分为分面组,并确保即使在分面之后树状图也能正确对齐。
创建于 2024 年 7 月 17 日,使用 reprex v2.1.0
~
可以从 ComplexHeatmap 创建更复杂的热图示例:
If you want to use
ggplot2to create a heatmap plot with dendrogram. I have created a package named ggalign, you can use it to create a heatmap as complex asComplexheatmap:Specifically, it can split the heatmap into facet groups and ensure the proper alignment of the dendrogram even after faceting.
Created on 2024-07-17 with reprex v2.1.0

~
A more complex heatmap example from the ComplexHeatmap can be created:
编辑
从 2011 年 8 月 8 日起,
ggdendro软件包可在 CRAN另请注意,树状图提取函数现在称为
dendro_data而不是cluster_data是的,确实如此。但目前您必须克服一些困难:
代码:
首先加载库并设置 ggplot 的数据:
提取树状图数据并创建绘图
使用网格图形和一些手动对齐来定位页面上的三个图
EDIT
From 8 August 2011 the
ggdendropackage is available on CRANNote also that the dendrogram extraction function is now called
dendro_datainstead ofcluster_dataYes, it is. But for the time being you will have to jump through a few hoops:
ggdendropackage (available from CRAN). This package will extract the cluster information from several types of cluster methods (includingHclustanddendrogram) with the express purpose of plotting inggplot.The code:
First load the libraries and set up the data for ggplot:
Extract dendrogram data and create the plots
Use grid graphics and some manual alignment to position the three plots on the page